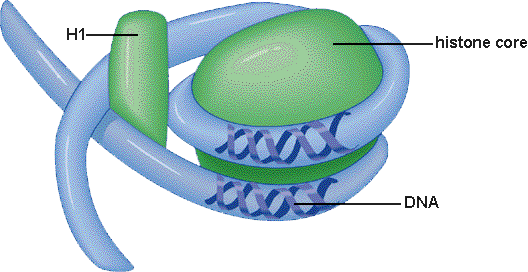
Limited to the facts that a nucleosome consists of 8 small histone protein molecules wrapped around with DNA and held together by another histone protein.
Nucleic acids and proteins (AHL)
8.1 DNA structure
8.1.1 Outline the structure of nucleosomes including histone proteins and DNA.
Limited to the facts that a nucleosome consists of 8 small histone protein molecules wrapped around with DNA and held together by another histone protein.


8.1.2 State that only a small proportion of the DNA in the nucleus constitutes genes and that the majority consists of repetitive sequences (cross reference 8.3.4).
The function of the repetitive sequences is not required, but students should know that their presence is made use of in profiling.
Major and minor grooves, direction of the ‘twist’, alternative B and Z forms, and details of the dimensions are not required.
8.2 DNA replication
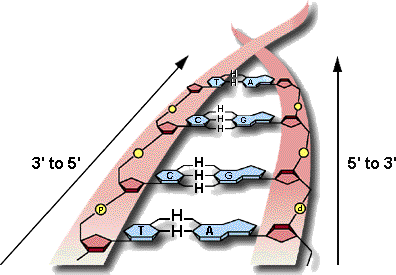
8.2.1 State that DNA replication occurs in a 5’ à 3’ direction.
8.2.2 Explain the process of DNA replication in eukaryotes including the role of enzymes (helicase, DNA polymerase III, RNA primase, DNA polymerase I and DNA ligase), Okazaki fragments and deoxynucleoside triphosphates.
Details of Meselson and Stahl’s experiment is not required. The function of the mentioned enzymes should be stated in general terms only. The explanation of Okasaki fragments in relation to the direction of action of DNA Polymerase III is required.
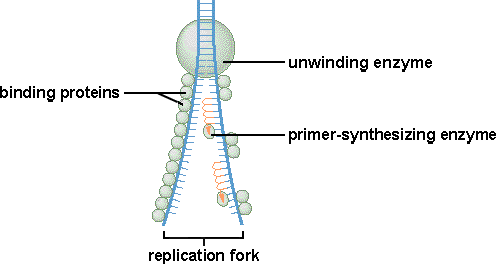
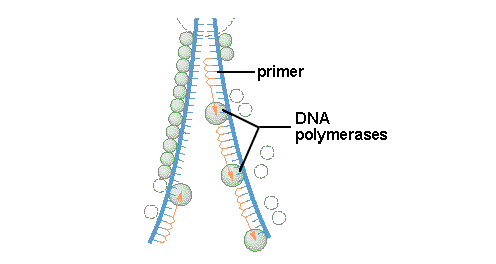

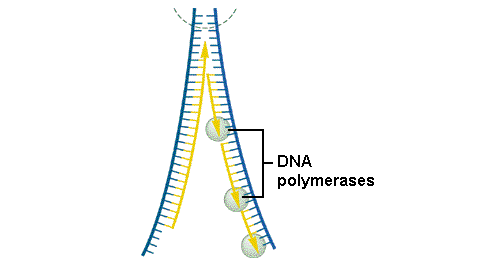
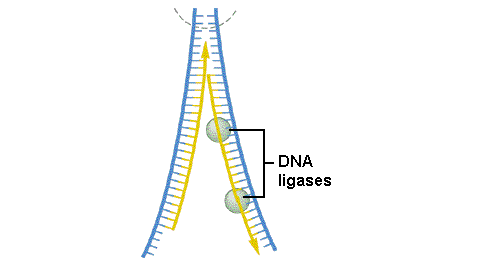
8.2.3 State that in an eukaryotic chromosome, replication is initiated at many points.
8.3 Transcription
8.3.1 State that transcription is carried out in a 5’ à 3’ direction.
8.3.2 Outline the Lac Operon model as an example of the control of gene expression in prokaryotes.
Mention that operons are found only in prokaryotes. Mention only the idea of a regulator gene producing a protein that prevents RNA polymerase binding to the promoter region.
 Check out this link
for a superb Lac Operon Model Demonstration
Check out this link
for a superb Lac Operon Model Demonstration
8.3.3 Explain the process of transcription in eukaryotes including the role of promoter region, RNA polymerase, ATP, and terminator.
The following details are not required: there is more than one type or RNA polymerase, features of the promoter region, the need for transcription protein factors for RNA polymerase binding, TATA boxes (and other repetitive sequences), the exact sequence of the bases which act as terminators.
Gene regulation can be limited to the presence of other genes (often on other chromosomes) that affect binding of RNA polymerase to the promoter region, and to the control of both the post-transcriptional modification of RNA and post-translational modification of proteins.
 Check out this link
for a superb demonstration of the process of Transcription
Check out this link
for a superb demonstration of the process of Transcription
8.3.4 State that eukaryotic chromosomes contain far more DNA than is needed to code for their protein products.
8.3.5 Outline the difference between introns and exons.
8.3.6 State that eukaryotic RNA needs the removal of introns to form mature mRNA and that this process is called splicing.
8.3.4- Further details of the process of post-transcriptional modification of RNA, mention of the heterogeneous nuclear RNA or the reasons for so many introns are not required.
 Check out this link
for a superb demonstration of the process of Splicing
Check out this link
for a superb demonstration of the process of Splicing
8.3.7 State that a small group of viruses, known as retroviruses, cause host cells to synthesise viral reverse transcriptase.
8.3.8 State that reverse transcriptase catalyses the production of single-stranded ‘novel’ DNA from RNA.
8.3.9 Explain why reverse transcriptase is a useful tool for molecular biologists.Here is an opportunity to relate some aspects of the DNA viral life cycle with that of the AIDS virus (an RNA virus, albeit an unusual one!). Viral RNA being transcribed into DNA that forms part of the genome of a lymphocyte nucleus. The latter cell divides, copying the ‘viral DNA’. After dormancy the lymphocyte manufactures the viral mRNA and many virus particles are made.
8.4 TranslationThis enzyme can make DNA from mature mRNA (e.g., of human insulin), which can then be spliced into host DNA (e.g., E. coli), without the introns.
8.4.1 Outline that the structure of a tRNA allows recognition by a tRNA activating enzyme that binds a specific amino acid to it using ATP for energy.
Each amino acid has a specific tRNA activating enzyme (the name aminoacyl-tRNA synthetase is not required). Degeneracy, that is, some amino acids having more than one tRNA, should also be included.
8.4.2 Outline the structure of ribosomes including protein and RNA composition, large and small subunits, two tRNA binding sites and mRNA binding sites.
The ‘coverleaf’ shape of tRNA and CCA at the 3’ end should be included. Mention that prokaryote, chloroplast and mitochondrial ribosomes are 70s, whereas eukaryote ribosomes are 80s. Knowledge of the derivation of Svedberg units is not required.
8.4.3 State that translation consists of initiation, elongation and termination.
8.4.4 State that translation occurs in a 5’ à 3’ direction.
8.4.5 Explain in detail the process of translation including GTP, ribosomes (including peptidyl transferase), polysomes, start codon and stop codons.
Mention of the P and A sites, initiating methione, T factor and recall of actual stop codons are not required.
 Check out this link
for a superb demonstration of the process of Translation
Check out this link
for a superb demonstration of the process of Translation
8.4.6 State that free ribosomes synthesise proteins for use primarily within the cell itself and that bound ribosomes synthesise proteins primarily for secretion and lysosomes.
8.5 Proteins
8.5.1 Explain the four levels of structure of proteins, indicating their significance.
Primary: linear sequence of amino acids with peptide linkages. (Regard disulfide bridges as part of the tertiary structure). Names of specific amino acids, mention of L- and D- forms or branching vs. unbranched, are not required, although mention of R side chains leading to polar/non-polar amino acids is needed for 8.5.2. Note that changes in the sequence may have several effects on the overall structure and activity. Mention the almost infinite number of sequences. Secondary: the formation of the a -helix (e.g., keratin – hair, wool, horn, feathers, etc.) and b -pleated sheets (e.g., silk), held together by hydrogen bonds. Tertiary: mention the possibility of creating ‘active sites’, added strength due to ionic bonds and disulfide bridges and the possibility of prosthetic groups and coenzymes. Quaternary: note that most large, non-structural proteins have more than one polypeptide and that it leads to greater range of biological activity.
8.5.2 Outline the difference between fibrous and globular proteins, with reference to two examples of each type.
8.5.3 Explain the significance of polar and non-polar amino acids (cross reference 7.1.3, 1.4.1 and 1.4.2).
Limited to polar amino acids leading to proteins with greater hydrophilic tendencies and water solubility due to the hydrogen bond formation; the reverse being true for proteins rich in non-polar amino acids. This should be related to how parts of large globular proteins interact with membranes.
8.5.4 State six functions of proteins, giving a named example of each.
8.6 Enzymes
8.6.1 State that metabolic pathways consist of chains and cycles of enzyme catalysed reactions.
8.6.2 Describe the "induced fit" model.
An extension of the ‘lock-and-key’ model. Its importance in the reduction of the activation energy should be mentioned and how it can account for the broad specificity of some enzymes (the ability to bind several substrates).
8.6.3 Explain that enzymes lower the activation energy of the chemical reactions that they catalyse.
Graphical representation of both exergonic and endergonic reactions should be covered, but no specific energy values need be recalled. An understanding how to calculate the activation energy of a reaction when reversed is also needed.
Competitive: explained by an inhibiting molecule so similar to the substrate molecule that it binds to the active site so preventing the substrate binding. Examples: inhibition of butanedioic acid (succinate) dehydrogenase by propanedioic acid (malonate) in the Krebs cycle; sulfonamide Prontosil™ (an antibiotic) inhibits folic acid synthesis in bacteria.
Non-competitive: limited to an inhibitor molecule binding to an enzyme (not to its active site) that results in a confonnational change in its active site resulting in a change in activity. Examples include Hg2+, Ag+, Cu2+ and CN- ion inhibition of many enzymes (including those like cytochrome oxidase) by binding to –SH groups, thereby breaking –S-S- linkages; nerve gases like sarin and DFP inactivate ethanoyl (acetyl) cholinesterase; Lisinopril™ lowers high blood pressure by inhibiting the enzyme ACE which manufactures angiotensin II.
8.6.5 Explain the role of allostery with respect to feedback inhibition and the control of metabolic pathways.
Allostery as a form of non-competitive inhibition. Mention that all allosteric enzymes consist of two or more polypeptides whose shape can be altered by the binding of effectors to an allosteric state, thereby increasing or decreasing its activity. Metabolites can act as allosteric inhibitors of enzymes earlier in a metabolic pathway and regulate metabolism according to the requirements of organisms; a form of negative feedback. One example is ATP inhibition of phosphofructokinase (glycolysis); another is inhibition of aspartate carbamoyltransferase (ATCase) (which catalyses the first step in pyrimidine synthesis). ATP is a positive modulator and CTP a negative modulator. Graphical representation of allosteric enzyme action is not required.